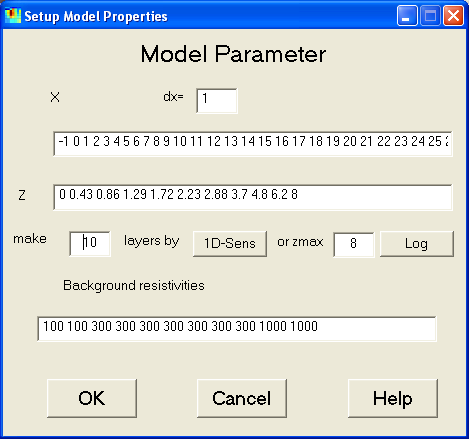
This dialog is used to control the model parameterizations.
By typing dx a new (equidistant) x-grid is defined, which also can be
edited manually.
The boundaries of the model layers can be changed both manually or automatically
(by regarding 1D sensitivities or by constructing logarithmically increasing
thicknesses for a given model depth).
Furthermore, the background resistivities can be set up. In the presented
example a three-layer case is introduced. Note, that the number of Background
resistivities must equal the number of z-Layers, since the last resistivity
is that below the model.